Find the imbalance of each node, that is the number of tip descendants of each of its two children. With thanks to Leonid Chindelevitch for use of code from computeLRValues from treeCentrality.
treeImb(tree)Arguments
Value
A matrix where rows correspond to nodes of the tree. The two column entries correspond to the number of tip descendants of each of its two children. (Note that this is the transform of the output in phyloTop version 1.0.0.) Where the row number corresponds to a tip, the entries are (0,0).
See also
Examples
## Find the imbalance numbers in a random tree with 10 tips:
tree <- rtree(10)
plot(tree)
nodelabels()
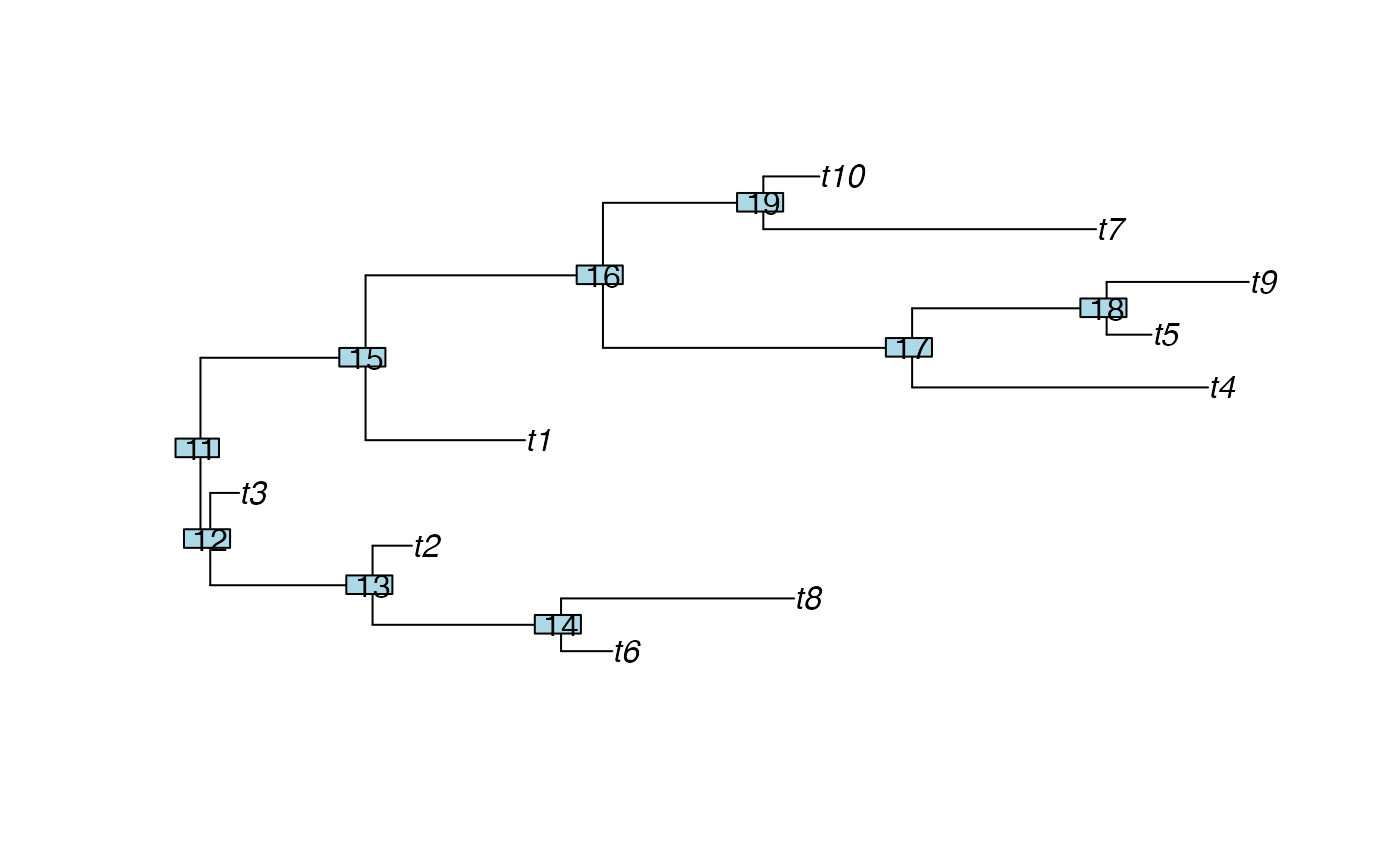 treeImb(tree)
#> [,1] [,2]
#> t1 0 0
#> t9 0 0
#> t8 0 0
#> t3 0 0
#> t6 0 0
#> t7 0 0
#> t2 0 0
#> t5 0 0
#> t10 0 0
#> t4 0 0
#> node11 4 6
#> node12 5 1
#> node13 2 3
#> node14 1 2
#> node15 1 1
#> node16 1 1
#> node17 1 3
#> node18 1 2
#> node19 1 1
treeImb(tree)
#> [,1] [,2]
#> t1 0 0
#> t9 0 0
#> t8 0 0
#> t3 0 0
#> t6 0 0
#> t7 0 0
#> t2 0 0
#> t5 0 0
#> t10 0 0
#> t4 0 0
#> node11 4 6
#> node12 5 1
#> node13 2 3
#> node14 1 2
#> node15 1 1
#> node16 1 1
#> node17 1 3
#> node18 1 2
#> node19 1 1