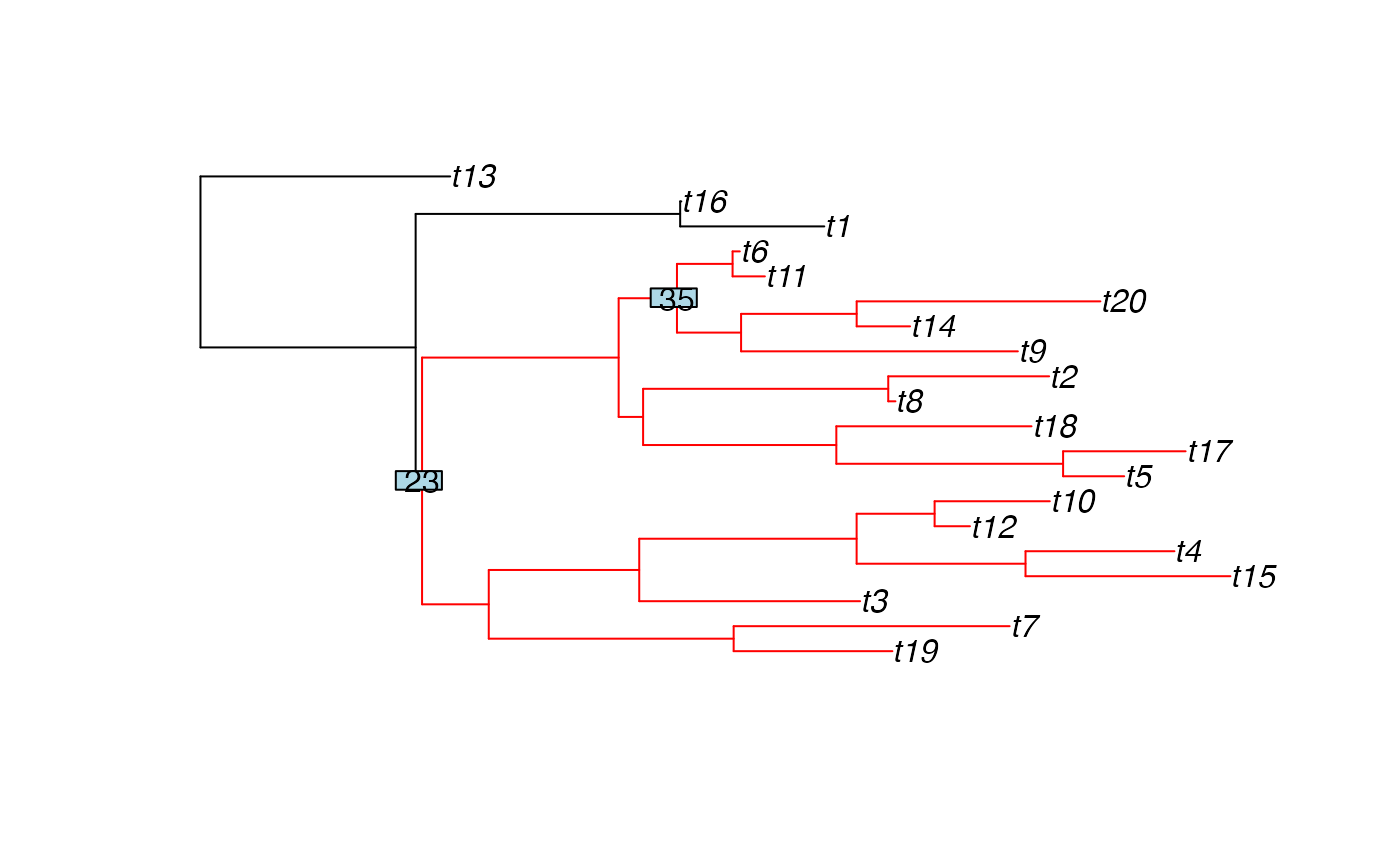
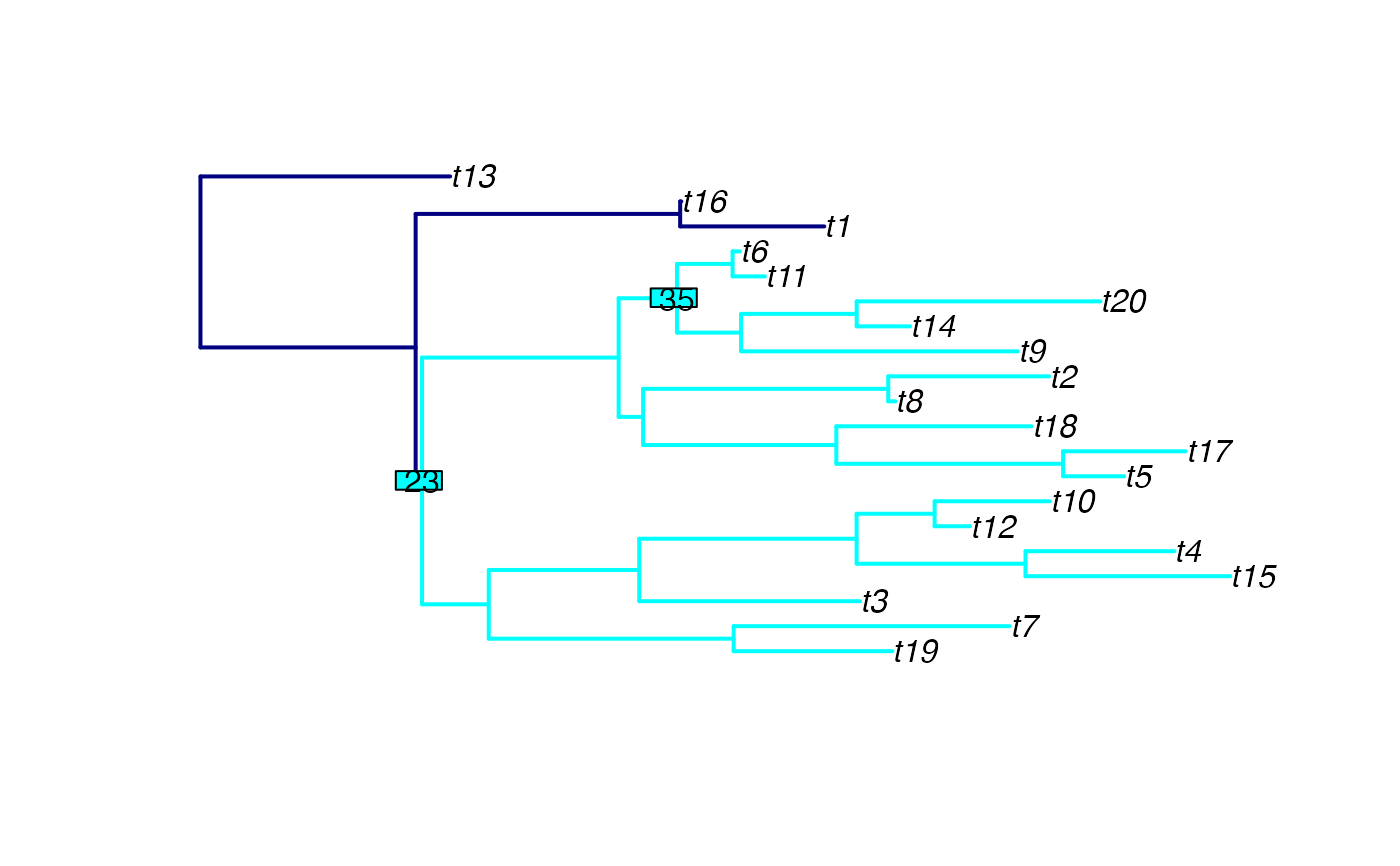
Plot a tree, highlighting the clade(s) descending from the given node(s)
subtreeShow(
tree,
nodeList,
showNodeLabels = "nodeList",
mainCol = "black",
subtreeCol = "red",
nodeLabelCol = "lightblue",
...
)Arguments
- tree
a tree of class
phyloorphylo4. The tree should be binary and rooted; if not it will be coerced into a binary rooted tree using multi2di, if possible.- nodeList
a list of one or more internal nodes in the tree.
- showNodeLabels
option of whether to show node labels. Default is "nodeList", which only labels the nodes in
nodeList. ChoosingshowNodeLabels="all"will display all node labels; any other arguments will remove all node labels.- mainCol
colour for the edges which are not highlighted (default is black).
- subtreeCol
colour for the edges which are highlighted (default is red).
- nodeLabelCol
background colour for any node labels shown (default is light blue)
- ...
further arguments to be passed to plot.phylo
Value
A plot of the tree, with clade(s) descending from the given node(s) highlighted.