Finds the sizes of configurations in the tree.
nConfig(tree, maxClade = NULL)Arguments
Value
A list with 2 entries:
cladeSizes is a vector giving the size of the clade descending at each node:tips all have the value 1, internal nodes have their number of tip descendants.
numClades is a vector where numClades[[i]] is the number of clades of size i in the tree. All clade sizes are calculated, but the output can be restricted using
maxCladeto just those of size up to 'maxClade'.
Examples
## Configuration sizes on a random tree with 10 tips:
tree <- rtree(10)
plot(tree)
nodelabels()
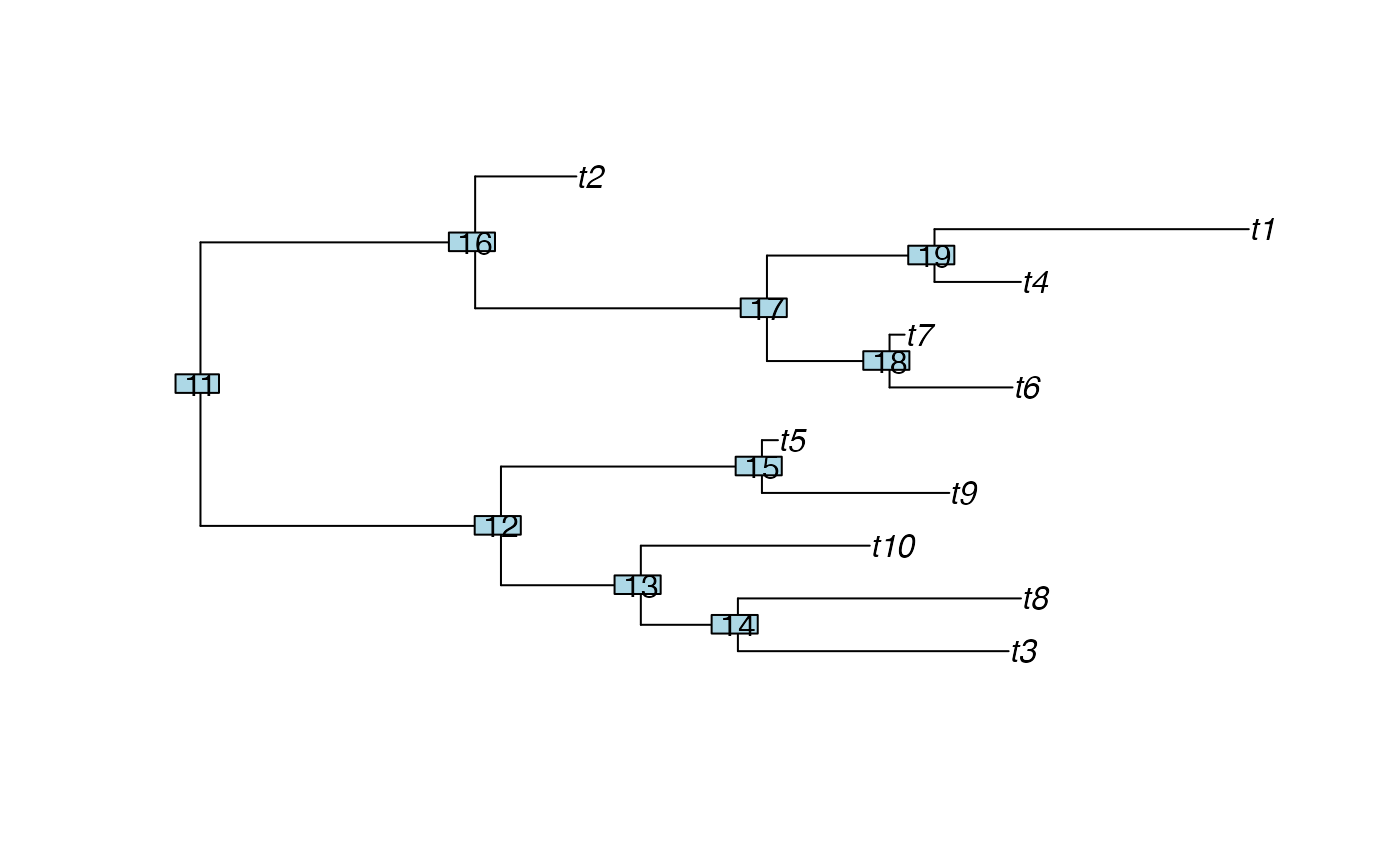 nConfig(tree)
#> $cladeSizes
#> t2 t1 t3 t5 t4 t10 t8 t7 t9 t6 node11
#> 1 1 1 1 1 1 1 1 1 1 10
#> node12 node13 node14 node15 node16 node17 node18 node19
#> 7 3 2 4 2 2 3 2
#>
#> $numClades
#> 1 2 3 4 5 6 7 8 9 10
#> 10 4 2 1 0 0 1 0 0 1
#>
nConfig(tree)
#> $cladeSizes
#> t2 t1 t3 t5 t4 t10 t8 t7 t9 t6 node11
#> 1 1 1 1 1 1 1 1 1 1 10
#> node12 node13 node14 node15 node16 node17 node18 node19
#> 7 3 2 4 2 2 3 2
#>
#> $numClades
#> 1 2 3 4 5 6 7 8 9 10
#> 10 4 2 1 0 0 1 0 0 1
#>